Overview:
Hello everyone, today I share an article "Spatiotemporal and direct capturing global substrates of lysine-modifying enzymes in living cell" published in Nature Communications. The corresponding author is Shanghai Pharmaceutical Research, Chinese Academy of Sciences. Researcher Chen Xiaohua of the Institute; the research group mainly conducts research on protein interactions and protein homeostasis regulation, combining multi-disciplinary approaches such as medicinal chemistry, organic chemistry, molecular and cell biology, protein engineering, chemical proteomics and a variety of emerging technologies. Carry out protein-protein interaction and target confirmation research, and develop innovative drugs based on targeted protein degradation technology.

Background:
Post-translational modification refers to the covalent chemical modification of amino acid residues that occurs after protein synthesis by ribosomes. Most of these post-translational modifications require the participation of enzymes. Post-translational modifications are very important for regulating protein function, and dysregulation of post-translational modifications can lead to a variety of complex diseases. Therefore, it becomes important to accurately characterize the association between post-translational modification-related enzymes and substrates. Currently, common analysis strategies are substrate-centered to identify the corresponding protein-modifying enzymes, such as chemical proteomics based on PTM-modified substrate surrogates, protein (peptide) microarrays, and co-immunoprecipitation. Current interaction analysis of enzymes and substrates is usually performed in vitro, which suffers from various shortcomings. For example, it is impossible to simulate the complex and dynamic native cellular environment and it is difficult to capture weakly and transiently interacting substrates. Based on the above considerations, the researchers developed a chemical cross-linking method that directly captures post-translational modification enzyme substrates in living cells, achieving global analysis of post-translational modification enzymes and direct analysis of modification sites.
Key data:
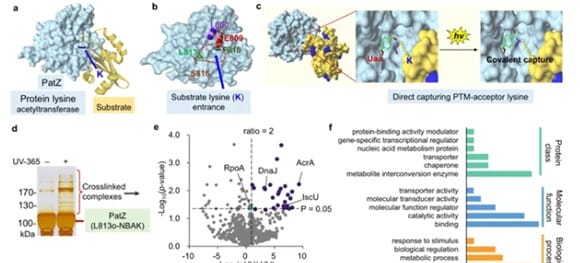
11. Capture strategy design for post-translational modification enzyme substrates
Chemical cross-linking technology is widely used in protein-protein interactions Inspired by this, the authors hope to directly analyze enzyme substrates in living cells by integrating chemical cross-linking technology and covalent binding processes of post-translational modifications. In this article, the author chose acylation modification on protein lysine residues as the research object. The authors first used an unnatural amino acid (Uaa) o-nitrophenol-derived lysine (o-NBAK) as a photocrosslinker, which can specifically and efficiently bind to other lysines under light. By embedding o-NBAK in the catalytic pocket of the enzyme, acylated lysine residues on the interacting substrates will be captured in a spatially and temporally controlled manner with high sensitivity and precision.
2. Proteome characterization of the direct substrate of lysine acetyltransferase PatZ
The researchers analyzed the PatZ enzyme in Escherichia coli. PatZ and its homologous enzymes are the main lysine acetyltransferases in bacteria and regulate a variety of biological processes, including growth, motility, stress response, etc., but the corresponding acylation substrates remain elusive. Therefore, PatZ and its cognate enzymes display diverse acylation activities in addition to acetylation. To realize the substrate analysis of PatZ enzyme, we first need to find the appropriate site for introducing o-NBAK. Since there is currently no available crystal structure, the authors conducted homology modeling of the catalytic domain of PatZ. Three sites (I800, F810, L813) in PatZ were selected for the introduction of o-NBAK Uaa. It was found that the PatZ mutant introducing o-NBAK at the I800 and L813 sites can achieve good chemical cross-linking effects, while no chemical cross-linking of the protein was observed when o-NBAK Uaa was introduced at a position far away from the catalytic pocket (S816). This result demonstrates the spatial resolution characteristics of this strategy (Fig. 3). The authors then used mass spectrometry and WB to verify the identified potential substrates and acetylation sites of PatZ to prove the reliability of the results.
3. Applicability of photocatalytic chemical cross-linking method to capture post-translational modification enzyme substrates
Based on the successful analysis of PatZ substrates, the authors successfully applied this strategy to other enzymes, including YiaC in bacteria , LplA, TmcA, YjaB enzymes (Fig. 4) and Tip60 and GCN5 enzymes in mammals. The results show that although these enzymes are all acetylation-modifying enzymes, their direct substrates are very different.
Summary:
The strategy of directly capturing PTM-enzyme substrates provides a new chemical tool for global substrate analysis of PTM-enzymes in living cells, greatly expanding the scope of PTM-enzymes. Enzyme substrate profile. This strategy has a high degree of residue selectivity, spatial and temporal resolution, and can achieve covalent capture, making it possible to decode substrates with higher sensitivity and accuracy, while also directly verifying the acylation site of the substrate. The concept of direct capture of PTM-enzyme substrates may inspire and lead to the further development of advanced capture strategies based on other kinds of residue-selective cross-linking chemistries, expanding the study of other types of post-translational modifications.

 扫一扫微信交流
扫一扫微信交流
发布评论